Subset SWOT LR L2 Unsmoothed data from AVISO’s THREDDS Data Server
This notebooks explains how to retrieve a geographical subset of unsmoothed (250-m) SWOT LR L2 data on AVISO’s THREDDS Data Server.
L2 Unsmoothed data can be explored at:
You need to have xarray, numpy, pydap, threddsclient and matplotlib+cartopy (for visualisation) packages installed in your Python environment to execute this notebook.
Warning
Threddsclient must be patched with adding the OPeNDAP name in the nodes.OPENDAP_SERVICE list as : OPENDAP_SERVICE = [“OPENDAP”, “OpenDAP”, “OPeNDAP”]
Warning
This tutorial shows how to retrieve SWOT LR L2 Unsmoothed datasets via the Opendap protocol of the AVISO’s THREDDS Data Server, that can be time consuming for large datasets. If you need to download large number of L2 LR SSH Unsmoothed half orbits, please use the AVISO’s FTP as in this tutorial.
Tutorial Objectives
Request Aviso’s Thredds Data Server catalogue to find unsmoothed products with cycles/passes numbers
Subset data intersecting a geographical area
Download data locally and visualise it
Import + code
[1]:
# Install Cartopy with mamba to avoid discrepancies
# ! mamba install -q -c conda-forge cartopy
[2]:
import os
from getpass import getpass
import threddsclient
import numpy as np
import xarray as xr
from xarray.backends import PydapDataStore
import requests as rq
import cartopy.crs as ccrs
import matplotlib.pyplot as plt
[3]:
def retrieve_matching_datasets(url_catalogue, level, variant, cycle_min, cycle_max, half_orbits):
""" Returns the list of datasets available in the catalogue, matching required half_orbits and cycles range.
Args:
url_catalogue: the catalogue
level: swot LR data level ("L2", "L3")
variant: Swot LR data variant ("Basic", "Expert", "Unsmoothed", "WindWave")
cycle_min: minimum cycle
cycle_max: maximum cycle
half_orbits: list of passes numbers
Returns:
The list of matching dataset nodes.
"""
import re
def _swot_lr_filename_match(filename, level, variant, cycle_min, cycle_max, half_orbits):
pattern = f'SWOT_{level}_LR_SSH_{variant}'
match_object = re.match(pattern+r'_(\d+)_(\d+)_(.*)', filename)
cycle_number = int(match_object.group(1))
pass_number = int(match_object.group(2))
return pass_number in half_orbits and cycle_number >= cycle_min and cycle_number <= cycle_max
return list(filter(
lambda dataset: (
_swot_lr_filename_match(dataset.name, level, variant, cycle_min, cycle_max, half_orbits)),
threddsclient.crawl(url_catalogue, depth=2)))
def open_dataset(dataset_url):
""" Open the dataset at dataset_url.
Args:
dataset_url
Returns:
xr.Dataset
"""
session = rq.Session()
session.auth = (username, password)
try:
store = PydapDataStore.open(dataset_url, session=session, timeout=300, user_charset='UTF-8')
return xr.open_dataset(store)
except Exception as e:
print(f"Something wrong with opendap url {dataset_url}: {e}")
def _open_dataset_coords(dataset_url):
positions_url = dataset_url + '?' + ','.join(["/left/latitude", "/left/longitude", "/right/latitude", "/right/longitude"])
ds_coords = open_dataset(positions_url)
return ds_coords
def _get_indexes(ds, lon_range, lat_range):
mask_left = ((ds["/left/longitude"] >= lon_range[0]) & (ds["/left/longitude"] <= lon_range[1]) &
(ds["/left/latitude"] >= lat_range[0]) & (ds["/left/latitude"] <= lat_range[1]))
mask_right = ((ds["/right/longitude"] >= lon_range[0]) & (ds["/right/longitude"] <= lon_range[1]) &
(ds["/right/latitude"] >= lat_range[0]) & (ds["/right/latitude"] <= lat_range[1]))
mask = (mask_left | mask_right).any('num_pixels')
return np.where(mask)[0][0], np.where(mask)[0][-1]
def _build_dataset_groups(ds, left_vars, right_vars, attrs):
# Clean group EXTRA_DIMENSION and variable attributes from global attributes
attributes = dict(filter(
lambda item: (
item[0] not in ["_NCProperties"] and
f"/left/" not in item[0] and
f"/right/" not in item[0] and
"EXTRA_DIMENSION" not in item[0]),
attrs.items()))
ds_left = ds[left_vars]
ds_left.attrs.clear()
ds_left.attrs["description"] = attributes.pop("left_description")
ds_left = ds_left.rename({v: v.removeprefix('/left/') for v in left_vars})
for var in list(ds_left.keys()):
ds_left[var].encoding = {'zlib':True, 'complevel':5}
ds_right = ds[right_vars]
ds_right.attrs.clear()
ds_right.attrs["description"] = attributes.pop("right_description")
ds_right = ds_right.rename({v: v.removeprefix('/right/') for v in right_vars})
for var in list(ds_right.keys()):
ds_right[var].encoding = {'zlib':True, 'complevel':5}
ds_root = ds[[]]
ds_root.attrs = attributes
return ds_root, ds_left, ds_right
def to_netcdf(ds_root, ds_left, ds_right, output_file):
""" Writes the dataset to a netcdf file """
ds_root.to_netcdf(output_file)
ds_right.to_netcdf(output_file, group='right', mode='a')
ds_left.to_netcdf(output_file, group='left', mode='a')
def _output_dir_prompt():
answer = input('Do you want to write results to Netcdf files? [y/n]')
if not answer or answer[0].lower() != 'y':
return None
return input('Enter existing directory:')
def load_subsets(matching_datasets, variables, lon_range, lat_range, output_dir=None):
""" Loads subsets with variables and lon/lat range, and eventually write them to disk.
Args:
matching_datasets : the datasets nodes in the catalogue
variables : the variables names
lon_range : the longitude range
lat_range : the latitude range
output_dir : the output directory to write the datasets in separated netcdf files
Returns:
xr.Dataset
"""
if not output_dir:
output_dir = _output_dir_prompt()
left_vars = [f'/left/{v}' for v in variables]
right_vars = [f'/right/{v}' for v in variables]
return [dataset for dataset in [_load_subset(dataset, left_vars, right_vars, lon_range, lat_range, output_dir) for dataset in matching_datasets] if dataset is not None]
def _load_subset(dataset_node, left_vars, right_vars, lon_range, lat_range, output_dir=None):
if output_dir:
output_file = os.path.join(output_dir, f"subset_{dataset_node.name}")
if os.path.exists(output_file):
print(f"Subset {dataset_node.name} already exists. Reading it...")
return xr.open_dataset(output_file)
# Open the dataset only with coordinates
dataset_url = dataset_node.opendap_url()
print(dataset_node.opendap_url())
ds_positions = _open_dataset_coords(dataset_url)
# Locate indexes of lines matching with the required geographical area
try:
idx_first, idx_last = _get_indexes(ds_positions, lon_range, lat_range)
except IndexError:
return None
# Download subset
dataset = open_dataset(dataset_url)
ds = xr.merge([dataset[var][idx_first:idx_last] for var in left_vars+right_vars])
print(f"Load intersection with selected area in dataset {dataset_node.name}: indices ({idx_first}, {idx_last})")
ds.load()
if output_file:
ds_root, ds_left, ds_right = _build_dataset_groups(ds, left_vars, right_vars, ds_positions.attrs)
to_netcdf(ds_root, ds_left, ds_right, output_file)
print(f"File {output_file} created.")
print('\n')
return ds
def _interpolate_coords(ds):
lon = ds.longitude
lat = ds.latitude
shape = lon.shape
lon = np.array(lon).ravel()
lat = np.array(lat).ravel()
dss = xr.Dataset({
'longitude': xr.DataArray(
data = lon,
dims = ['time']
),
'latitude': xr.DataArray(
data = lat,
dims = ['time'],
)
},)
dss_interp = dss.interpolate_na(dim="time", method="linear", fill_value="extrapolate")
ds['longitude'] = (('num_lines', 'num_pixels'), dss_interp.longitude.values.reshape(shape))
ds['latitude'] = (('num_lines', 'num_pixels'), dss_interp.latitude.values.reshape(shape))
def normalize_coordinates(ds):
""" Normalizes the coordinates of the dataset : interpolates Nan values in lon/lat, and assign lon/lat as coordinates.
Args:
ds: the dataset
Returns:
xr.Dataset: the normalized dataset
"""
_interpolate_coords(ds)
return ds.assign_coords(
{"longitude": ds.longitude, "latitude": ds.latitude}
)
Parameters
Authentication parameters
Enter your AVISO+ credentials
[4]:
username = input("Enter username:")
Enter username: aviso-swot@altimetry.fr
[5]:
password = getpass(f"Enter password for {username}:")
Enter password for aviso-swot@altimetry.fr: ········
Data parameters
Define the variables you want:
[7]:
variables = ['latitude', 'longitude', 'time', 'sig0_karin_2', 'ancillary_surface_classification_flag', 'ssh_karin_2', 'ssh_karin_2_qual']
OPTIONAL Define existing output folder to save results:
[8]:
output_dir = "downloads"#or None
Define the url of the catalog from which you want to extract data
[9]:
url_catalogue="https://tds-odatis.aviso.altimetry.fr/thredds/catalog/dataset-l2-swot-karin-lr-ssh-validated/PIC2/Unsmoothed/catalog.html"
level = 'L2'
variant = 'Unsmoothed'
Define the parameters needed to retrieve data:
geographical area
phase: 1day-orbit (Calval) / 21day-orbit (Science)
cycle min, max
list of half-orbits
Note
Passes matching a geographical area and period can be found using this tutorial
[10]:
# California
lat_range = 35, 42
lon_range = 233, 239
#phase, cycle_min, cycle_max = "calval", 400, 600
phase, cycle_min, cycle_max = "science", 23, 25
half_orbits = [11, 24, 317]
Area extraction
Gather datasets in the provided catalogue, matching the required cycles and half_orbits
[11]:
matching_datasets = retrieve_matching_datasets(url_catalogue, level, variant, cycle_min, cycle_max, half_orbits)
'num datasets =', len(matching_datasets)
[11]:
('num datasets =', 9)
[12]:
matching_datasets
[12]:
[<Node name: SWOT_L2_LR_SSH_Unsmoothed_023_011_20241022T144002_20241022T153050_PIC2_01.nc, content type: application/netcdf>,
<Node name: SWOT_L2_LR_SSH_Unsmoothed_023_024_20241023T014851_20241023T024018_PIC2_01.nc, content type: application/netcdf>,
<Node name: SWOT_L2_LR_SSH_Unsmoothed_023_317_20241102T130249_20241102T135416_PIC2_01.nc, content type: application/netcdf>,
<Node name: SWOT_L2_LR_SSH_Unsmoothed_024_011_20241112T112506_20241112T121554_PIC2_01.nc, content type: application/netcdf>,
<Node name: SWOT_L2_LR_SSH_Unsmoothed_024_024_20241112T223355_20241112T232522_PIC2_01.nc, content type: application/netcdf>,
<Node name: SWOT_L2_LR_SSH_Unsmoothed_024_317_20241123T094755_20241123T103922_PIC2_01.nc, content type: application/netcdf>,
<Node name: SWOT_L2_LR_SSH_Unsmoothed_025_011_20241203T081011_20241203T090059_PIC2_01.nc, content type: application/netcdf>,
<Node name: SWOT_L2_LR_SSH_Unsmoothed_025_024_20241203T191900_20241203T201026_PIC2_01.nc, content type: application/netcdf>,
<Node name: SWOT_L2_LR_SSH_Unsmoothed_025_317_20241214T063259_20241214T072426_PIC2_01.nc, content type: application/netcdf>]
[15]:
matching_datasets[0].opendap_url()
[15]:
'https://tds-odatis.aviso.altimetry.fr/thredds/dodsC/dataset-l2-swot-karin-lr-ssh-validated/PIC2/Unsmoothed/cycle_023/SWOT_L2_LR_SSH_Unsmoothed_023_011_20241022T144002_20241022T153050_PIC2_01.nc'
Subset data in the required geographical area
Warning
This operation may take some time : for each dataset, it downloads coordinates, calculates indices, loads subset and eventually writes it to netcdf file.
Set output_dir to None if you don’t want to write subsets to netcdf files.
[16]:
datasets_subsets = load_subsets(matching_datasets, variables, lon_range, lat_range, output_dir)
https://tds-odatis.aviso.altimetry.fr/thredds/dodsC/dataset-l2-swot-karin-lr-ssh-validated/PIC2/Unsmoothed/cycle_023/SWOT_L2_LR_SSH_Unsmoothed_023_011_20241022T144002_20241022T153050_PIC2_01.nc
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
Load intersection with selected area in dataset SWOT_L2_LR_SSH_Unsmoothed_023_011_20241022T144002_20241022T153050_PIC2_01.nc: indices (57896, 61318)
File downloads/subset_SWOT_L2_LR_SSH_Unsmoothed_023_011_20241022T144002_20241022T153050_PIC2_01.nc created.
https://tds-odatis.aviso.altimetry.fr/thredds/dodsC/dataset-l2-swot-karin-lr-ssh-validated/PIC2/Unsmoothed/cycle_023/SWOT_L2_LR_SSH_Unsmoothed_023_024_20241023T014851_20241023T024018_PIC2_01.nc
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
Load intersection with selected area in dataset SWOT_L2_LR_SSH_Unsmoothed_023_024_20241023T014851_20241023T024018_PIC2_01.nc: indices (21552, 25041)
File downloads/subset_SWOT_L2_LR_SSH_Unsmoothed_023_024_20241023T014851_20241023T024018_PIC2_01.nc created.
https://tds-odatis.aviso.altimetry.fr/thredds/dodsC/dataset-l2-swot-karin-lr-ssh-validated/PIC2/Unsmoothed/cycle_023/SWOT_L2_LR_SSH_Unsmoothed_023_317_20241102T130249_20241102T135416_PIC2_01.nc
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
Load intersection with selected area in dataset SWOT_L2_LR_SSH_Unsmoothed_023_317_20241102T130249_20241102T135416_PIC2_01.nc: indices (57902, 61397)
File downloads/subset_SWOT_L2_LR_SSH_Unsmoothed_023_317_20241102T130249_20241102T135416_PIC2_01.nc created.
https://tds-odatis.aviso.altimetry.fr/thredds/dodsC/dataset-l2-swot-karin-lr-ssh-validated/PIC2/Unsmoothed/cycle_024/SWOT_L2_LR_SSH_Unsmoothed_024_011_20241112T112506_20241112T121554_PIC2_01.nc
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
Load intersection with selected area in dataset SWOT_L2_LR_SSH_Unsmoothed_024_011_20241112T112506_20241112T121554_PIC2_01.nc: indices (57901, 61322)
File downloads/subset_SWOT_L2_LR_SSH_Unsmoothed_024_011_20241112T112506_20241112T121554_PIC2_01.nc created.
https://tds-odatis.aviso.altimetry.fr/thredds/dodsC/dataset-l2-swot-karin-lr-ssh-validated/PIC2/Unsmoothed/cycle_024/SWOT_L2_LR_SSH_Unsmoothed_024_024_20241112T223355_20241112T232522_PIC2_01.nc
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
Load intersection with selected area in dataset SWOT_L2_LR_SSH_Unsmoothed_024_024_20241112T223355_20241112T232522_PIC2_01.nc: indices (21548, 25037)
File downloads/subset_SWOT_L2_LR_SSH_Unsmoothed_024_024_20241112T223355_20241112T232522_PIC2_01.nc created.
https://tds-odatis.aviso.altimetry.fr/thredds/dodsC/dataset-l2-swot-karin-lr-ssh-validated/PIC2/Unsmoothed/cycle_024/SWOT_L2_LR_SSH_Unsmoothed_024_317_20241123T094755_20241123T103922_PIC2_01.nc
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
Load intersection with selected area in dataset SWOT_L2_LR_SSH_Unsmoothed_024_317_20241123T094755_20241123T103922_PIC2_01.nc: indices (57900, 61394)
File downloads/subset_SWOT_L2_LR_SSH_Unsmoothed_024_317_20241123T094755_20241123T103922_PIC2_01.nc created.
https://tds-odatis.aviso.altimetry.fr/thredds/dodsC/dataset-l2-swot-karin-lr-ssh-validated/PIC2/Unsmoothed/cycle_025/SWOT_L2_LR_SSH_Unsmoothed_025_011_20241203T081011_20241203T090059_PIC2_01.nc
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
Load intersection with selected area in dataset SWOT_L2_LR_SSH_Unsmoothed_025_011_20241203T081011_20241203T090059_PIC2_01.nc: indices (57895, 61315)
File downloads/subset_SWOT_L2_LR_SSH_Unsmoothed_025_011_20241203T081011_20241203T090059_PIC2_01.nc created.
https://tds-odatis.aviso.altimetry.fr/thredds/dodsC/dataset-l2-swot-karin-lr-ssh-validated/PIC2/Unsmoothed/cycle_025/SWOT_L2_LR_SSH_Unsmoothed_025_024_20241203T191900_20241203T201026_PIC2_01.nc
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
Load intersection with selected area in dataset SWOT_L2_LR_SSH_Unsmoothed_025_024_20241203T191900_20241203T201026_PIC2_01.nc: indices (21547, 25035)
File downloads/subset_SWOT_L2_LR_SSH_Unsmoothed_025_024_20241203T191900_20241203T201026_PIC2_01.nc created.
https://tds-odatis.aviso.altimetry.fr/thredds/dodsC/dataset-l2-swot-karin-lr-ssh-validated/PIC2/Unsmoothed/cycle_025/SWOT_L2_LR_SSH_Unsmoothed_025_317_20241214T063259_20241214T072426_PIC2_01.nc
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
/home/atonneau/.conda/envs/PY311_DEV/lib/python3.11/site-packages/pydap/handlers/dap.py:134: UserWarning: PyDAP was unable to determine the DAP protocol defaulting to DAP2. DAP2 is consider legacy and may result in slower responses.
Consider replacing `http` in your `url` with either `dap2` or `dap4` to specify the DAP protocol (e.g. `dap2://<data_url>` or `dap4://<data_url>`). For more
information, go to https://www.opendap.org/faq-page.
warnings.warn(
Load intersection with selected area in dataset SWOT_L2_LR_SSH_Unsmoothed_025_317_20241214T063259_20241214T072426_PIC2_01.nc: indices (57893, 61386)
File downloads/subset_SWOT_L2_LR_SSH_Unsmoothed_025_317_20241214T063259_20241214T072426_PIC2_01.nc created.
Basic manipulations
Concatenate subsets
ds = xr.concat(datasets_subsets, dim='num_lines') dsVisualize data on a pass
Open a pass dataset
[17]:
subset_file = "downloads/subset_SWOT_L2_LR_SSH_Unsmoothed_024_317_20241123T094755_20241123T103922_PIC2_01.nc"
[18]:
ds_left = xr.open_dataset(subset_file, group="left")
[19]:
ds_right = xr.open_dataset(subset_file, group="right")
Interpolate coordinates to fill Nan values in latitude and longitude, and assign them as coordinates.
[20]:
ds_left = normalize_coordinates(ds_left)
ds_right = normalize_coordinates(ds_right)
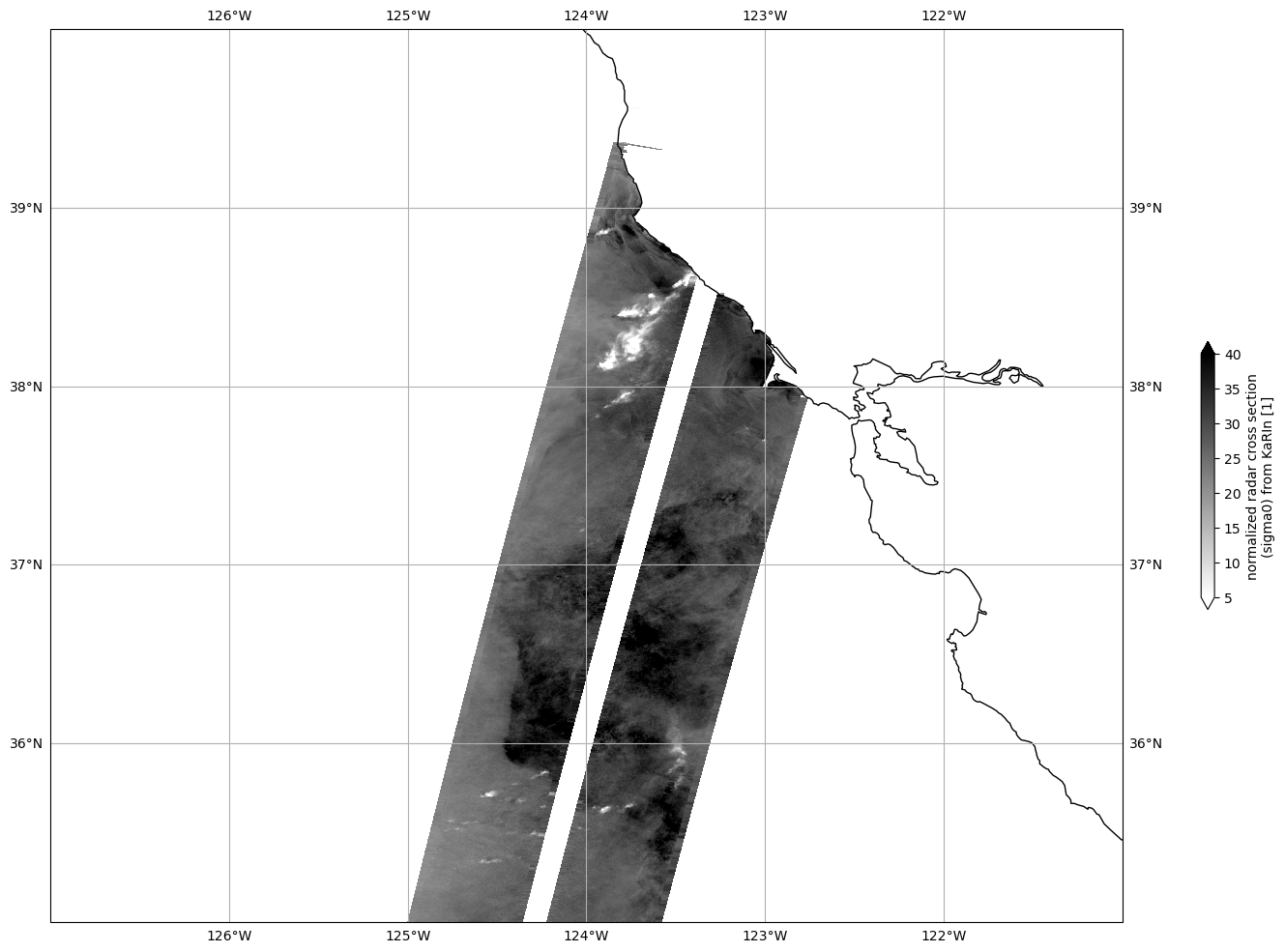
Plot Sigma 0
Mask invalid data
[21]:
for dss in ds_left, ds_right:
dss["sig0_karin_2"] = dss.sig0_karin_2.where(dss.ancillary_surface_classification_flag==0)
dss["sig0_karin_2"] = dss.sig0_karin_2.where(dss.sig0_karin_2 < 1e6)
#dss["sig0_karin_2_log"] = 10*np.log10(dss["sig0_karin_2"])
Plot data
[25]:
plot_kwargs = dict(
x="longitude",
y="latitude",
cmap="gray_r",
vmin=5,
vmax=40,
)
fig, ax = plt.subplots(figsize=(21, 12), subplot_kw=dict(projection=ccrs.PlateCarree()))
ds_left.sig0_karin_2.plot.pcolormesh(ax=ax, cbar_kwargs={"shrink": 0.3}, **plot_kwargs)
ds_right.sig0_karin_2.plot.pcolormesh(ax=ax, add_colorbar=False, **plot_kwargs)
ax.gridlines(draw_labels=True)
ax.coastlines()
ax.set_extent([-127, -121, 35, 40], crs=ccrs.PlateCarree())
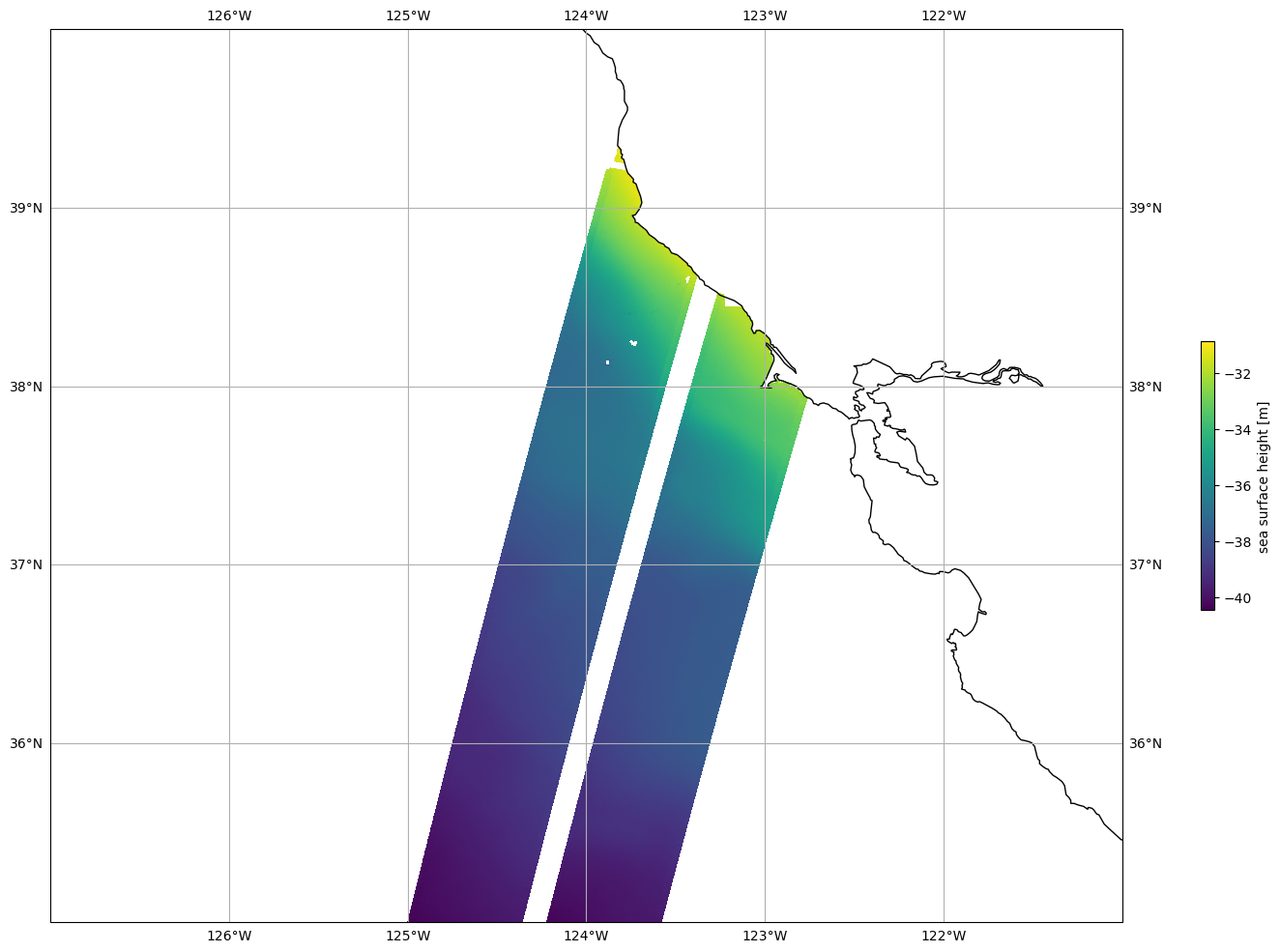
Plot SSHA
Mask invalid data
[23]:
for dss in ds_left, ds_right:
dss["ssh_karin_2"] = dss.ssh_karin_2.where(dss.ancillary_surface_classification_flag==0)
dss["ssh_karin_2"] = dss.ssh_karin_2.where(dss.ssh_karin_2_qual==0)
Plot data
[24]:
plot_kwargs = dict(
x="longitude",
y="latitude"
)
fig, ax = plt.subplots(figsize=(21, 12), subplot_kw=dict(projection=ccrs.PlateCarree()))
ds_left.ssh_karin_2.plot.pcolormesh(ax=ax, cbar_kwargs={"shrink": 0.3}, **plot_kwargs)
ds_right.ssh_karin_2.plot.pcolormesh(ax=ax, add_colorbar=False, **plot_kwargs)
ax.gridlines(draw_labels=True)
ax.coastlines()
ax.set_extent([-127, -121, 35, 40], crs=ccrs.PlateCarree())
[ ]:
